Testing Local Parallel Processing in CASA
Checking the output of parallelized tclean
CASA Parallelization Overview
Some tasks in CASA can be parallelized. NRAO claim the task that see the biggest gains in processing time are tclean, applycal, and flagdata.
Here we are going to test the tclean task with CASA 5.1.2. First I confirm that tclean produces the same results whether given a list of input files (TEST 1), or a single concatenated file (TEST 2). See the instructions here.
Initial conclusions:
■ As expected, tclean produces the same result whether fed individual files or a concatenated master file (using default options). However, the concatenated file has a shorter processing time.
■ Running tclean in parallel has spectacular time gains. The output looks very similar (but not identical), to the non-parallelized version.
■ The parallel output has marginally higher sum & mean pixel values. This increase only becomes significant when looking at the averaged spectrum. It does hold for regions of noise. It seems that paralleled tclean does a better job of recovering flux.
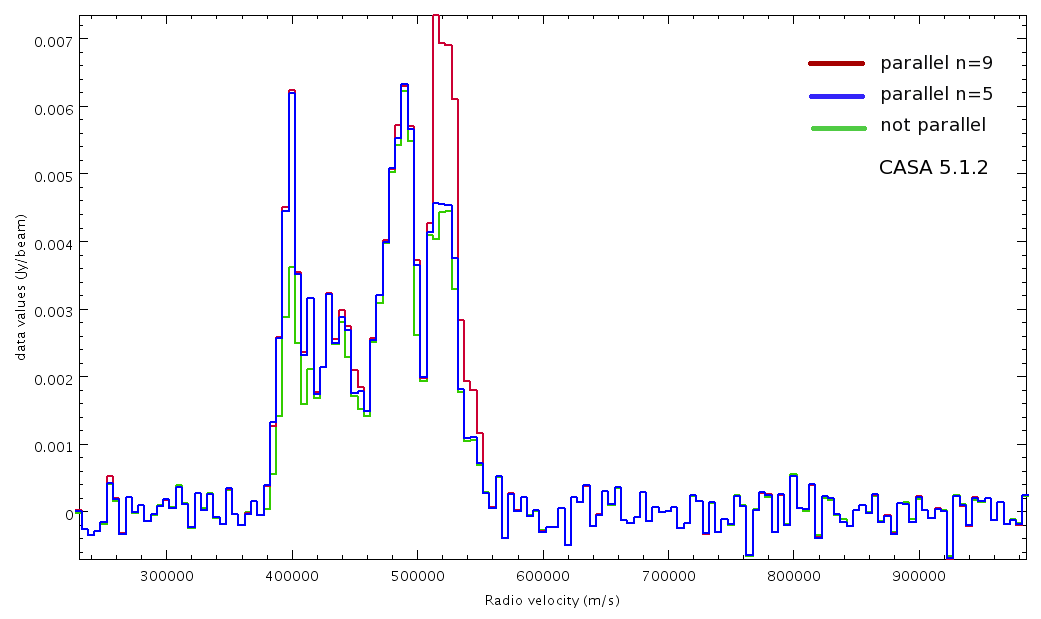
■ The averaged spectra at the bottom of the page show that these gains in flux are dependent on the number of cores chosen for processing. This has to do with how tclean generates an average spectra for each sub-MS.
NRAO expect parallel imaging to be signifcantly more stable from version 5.1.3 which should be released in May 2018. Tests will resume then.
TEST 1
Input: 10 separate measurement sets (from mosaic)
Parallel? No
Time: 3h45m
|
Full spectral cube:
Pixel sum : 91527
Pixel mean : 0.000147011
Standard deviation : 0.0752108
Skewness : -0.19447
Kurtosis : -3.05935
Minimum pixel value : -99.2058
At pixel : (47, 1723, 140, 1)
Maximum pixel value : 112.35
At pixel : (4, 1043, 26, 1)
Pixel median : -1.87703e-06
Total number of pixels : 622592000
Number of pixels used : 622585763 (100.0%)
No. of pixels excluded : 6237 (0.0%)
|
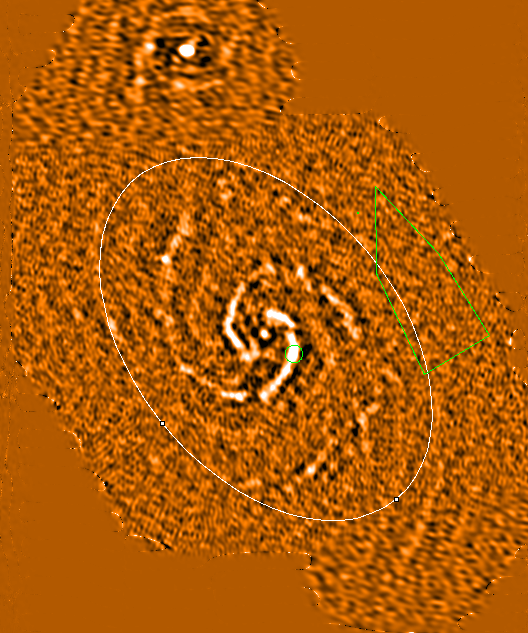
Ellipse of integrated intensity (see figure on right):
Pixel sum : 3.03969e+08
Pixel mean : 389.15
Standard deviation : 10265.4
Skewness : 2.06373
Kurtosis : -2.98812
Minimum pixel value : -37811.1
At pixel : (776, 1282, 1)
Maximum pixel value : 132687
At pixel : (887, 1222, 1)
Pixel median : -427.936
Total number of pixels : 4096000
Number of pixels used : 781109 (19.1%)
No. of pixels excluded : 3314891 (80.9%)
|
Integrated intensity (99.5% cut)
|
Histogram of ellipse data
|
Stats of noise region
(see polygon in figure)
Mean 0.00100928
Std Dev 0.0969997
Max 0.3325351
Min -0.3374273
Total 118.7307
|
Integrated intensity above 3-sigma:
Pixel sum : 6.89492e+08
Pixel mean : 40858.8
Standard deviation : 16799
Skewness : 2.10626
Kurtosis : 5.75119
Minimum pixel value : 25003
At pixel : (528, 1559, 1)
Maximum pixel value : 132687
At pixel : (887, 1222, 1)
Total number of pixels : 4096000
Number of pixels used : 16875 (0.4%)
No. of pixels excluded : 4079125 (99.6%)
|
TEST 2
Input: concat (with defaults) the 10 separate measurement sets into single file
Parallel? No
Time: 3h00m
|
Pixel sum : 90800
Pixel mean : 0.000145843
Standard deviation : 0.0752725
Skewness : -5.3911
Kurtosis : -2.97067
Minimum pixel value : -127.511
At pixel : (1581, 1136, 86, 1)
Maximum pixel value : 73.6167
At pixel : (43, 1425, 119, 1)
Pixel median : -1.87689e-06
Total number of pixels : 622592000
Number of pixels used : 622585773 (100.0%)
No. of pixels excluded : 6227 (0.0%)
|
Pixel sum : 3.03971e+08
Pixel mean : 389.153
Standard deviation : 10265.4
Skewness : 2.06374
Kurtosis : -2.98812
Minimum pixel value : -37810.3
At pixel : (776, 1282, 1)
Maximum pixel value : 132687
At pixel : (887, 1222, 1)
Pixel median : -428.004
Total number of pixels : 4096000
Number of pixels used : 781109 (19.1%)
No. of pixels excluded : 3314891 (80.9%)
|
|

|
Mean 0.001009395
Std Dev 0.09699952
Max 0.3325383
Min -0.3374223
Total 118.7442
|
Pixel sum : 6.89492e+08
Pixel mean : 40858.8
Standard deviation : 16799
Skewness : 2.10625
Kurtosis : 5.75115
Minimum pixel value : 25003.3
At pixel : (528, 1559, 1)
Maximum pixel value : 132687
At pixel : (887, 1222, 1)
Total number of pixels : 4096000
Number of pixels used : 16875 (0.4%)
No. of pixels excluded : 4079125 (99.6%)
|
TEST 3
Input: concat the 10 separate measurement sets into single file
Parallel? Yes
Cores: 9
Time: 45m
|
Pixel sum : 114150
Pixel mean : 0.000183347
Standard deviation : 0.0752537
Skewness : 7.72432
Kurtosis : -2.98198
Minimum pixel value : -60.1831
At pixel : (87, 1170, 53, 1)
Maximum pixel value : 86.9453
At pixel : (846, 2172, 94, 1)
Pixel median : -1.67351e-06
Total number of pixels : 622592000
Number of pixels used : 622585689 (100.0%)
No. of pixels excluded : 6311 (0.0%)
|
Pixel sum : 3.91518e+08
Pixel mean : 501.234
Standard deviation : 10287.3
Skewness : 2.14725
Kurtosis : -2.98803
Minimum pixel value : -37060.9
At pixel : (1075, 987, 1)
Maximum pixel value : 132870
At pixel : (888, 1221, 1)
Pixel median : -388.175
Total number of pixels : 4096000
Number of pixels used : 781109 (19.1%)
No. of pixels excluded : 3314891 (80.9%)
|
|
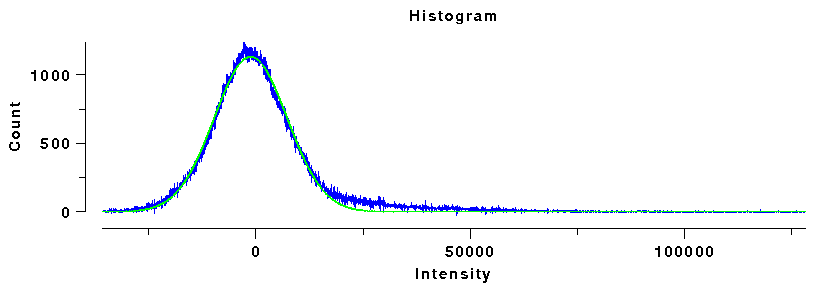
|
Mean 0.001009975
Std Dev 0.0969723
Max 0.3312833
Min -0.340028
Total 118.8124
|
Pixel sum : 7.21572e+08
Pixel mean : 40924
Standard deviation : 16735.3
Skewness : 2.07683
Kurtosis : 5.62795
Minimum pixel value : 25001.2
At pixel : (819, 1128, 1)
Maximum pixel value : 132870
At pixel : (888, 1221, 1)
Total number of pixels : 4096000
Number of pixels used : 17632 (0.4%)
No. of pixels excluded : 4078368 (99.6%)
|
TEST 4
Input: concat the 10 separate measurement sets into single file
Parallel? Yes
Cores: 5
Time: 57m
|
Pixel sum : 104136
Pixel mean : 0.000167264
Standard deviation : 0.0752265
Skewness : 7.57816
Kurtosis : -3.02897
Minimum pixel value : -74.4938
At pixel : (843, 2176, 59, 1)
Maximum pixel value : 104.372
At pixel : (1576, 1141, 137, 1)
Total number of pixels : 622592000
Number of pixels used : 622585810 (100.0%)
No. of pixels excluded : 6190 (0.0%)
Pixel sum : 3.45422e+08
Pixel mean : 442.22
Standard deviation : 10292.5
Skewness : 2.10265
Kurtosis : -3.01145
Minimum pixel value : -37141.9
At pixel : (895, 1169, 1)
Maximum pixel value : 132850
At pixel : (888, 1222, 1)
Total number of pixels : 4096000
Number of pixels used : 781109 (19.1%)
No. of pixels excluded : 3314891 (80.9%)
|
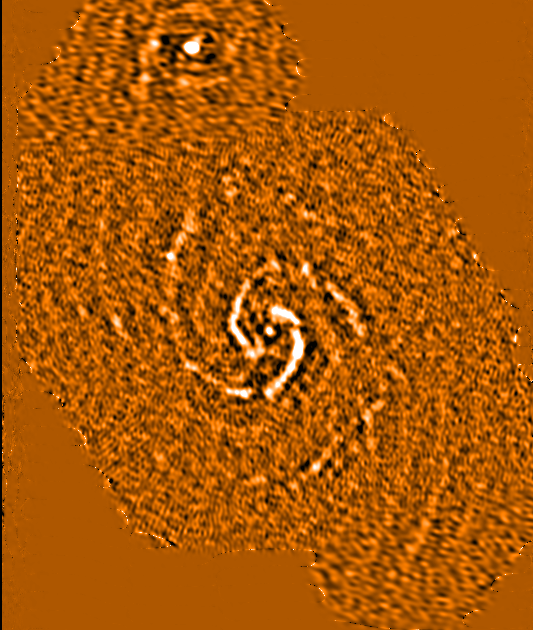
|
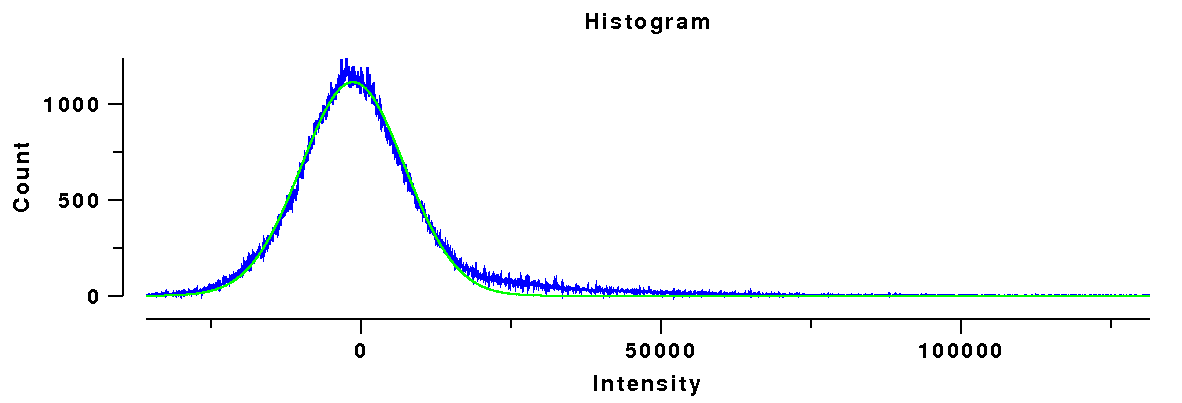
|
Mean 0.0009987468
Std Dev 0.09686029
Max 0.331913
Min -0.3406299
Total 117.4916
|
Pixel sum : 7.07172e+08
Pixel mean : 40891.2
Standard deviation : 16796.8
Skewness : 2.08016
Kurtosis : 5.62957
Minimum pixel value : 25000
At pixel : (702, 1317, 1)
Maximum pixel value : 132850
At pixel : (888, 1222, 1)
Total number of pixels : 4096000
Number of pixels used : 17294 (0.4%)
No. of pixels excluded : 4078706 (99.6%)
| |
On closer inspection, our tests show significantly different spectral profiles depending on the number of cores chosen.
The spectra plotted below show the average spectrum over a large circle enclosing most emission.
Differences are seen in the emission peaks, although the noise is identical. This difference arises from the degree to which CASA
splits the original dataset and the data it has available to determine an average spectrum.
CASA's viewer proved too slow to use with these data, so they were exported as fits and analyzed using Starlink. Example commands: