Using SMARechunker
SMARechunker can replace each of the SWARM chunks (called s1 through s4 in SWARM-only data sets, and s49 through s52 in mixed ASIC/SWARM datasets) with one or more chunks of reduced resolution and/or bandwidth.
SMA datasets consist of several files which reside in a single directory. SMARechunker reads the files in that input directory, and produces a new directory which contains the files required by the data reduction software, but with replacement SWARM chunks. No changes are made to the files in the input directory.
The syntax for SMARechunker is
$ SMARechunker [-d] -i {input data directory} -o {output data directory} [-r {n} reduce all SWARM chunks
by a factor of n}] [-f {first scan number}] [-l {last scan number}] {chunk:start channel:end channel:channels
to average}
Use -A for tracks which contain ASIC dataUse
-L to list the number of scans and chunks in the trackUse
-D 'ant1-ant2:rx:chunk:spikeLo-spikeHi:leftLo-leftHi:rightLo:rightHi' to despike (can be used more than once)
The most common thing to do, especially for continuum projects, is to use the -r option and reduce the resolution of all SWARM chunks by a constant factor. If more control is needed, one can specify the amount of averaging to be done for each chunk separately. So, for example
$ SMARechunker -i /sma/rtdata/science/mir_data/150701_07:26:14/ -o rebinned 49:0:16383:128 50:8000:8999:4
would read the data from /sma/rtdata/science/mir_data/150701_07:26:14/ and produce a new data set called 'rebinned' in your current working directory with SWARM chunk s49 (all 16384 channels) reduced in resolution by a factor of 128 (resulting in 16384/128 = 128 output channels) and SWARM chunk s50 channels 8000 to 8999 reduced in resolution by a factor of 4.
Remember that if your data set contains ASIC data, you must include the -A switch. If you are unsure about whether the dataset contains ASIC data, you can use the -L option to list all of the spectral chunks present in the dataset.
The optional -d switch tells the program to include the original full resolution and bandwidth chunks in the output data set..
The optional -D switch allows you to specify a spike which should be removed. The -D must be followed by a string, enclosed in quotes (to keep the shell from parsing it), with the following syntax:
-D 'a1-a2:rx:sb:chunk:spike1-spike2:left1-left2:right1-right2'
Where
a1 = antenna 1 (* = all) a2 = antenna 2 (* = all) rx = sideband (A or B) case ignored sb = sideband (u, l, or b) case ignored chunk = 1, 2, 3 or 4 spike1 = low channel number of spike spike2 = high channel number of spike left1 = low channel of left side for fit left2 = high channel of left side for fit right1 = low channel of right side for fit right2 = high channel of right side for fitMultiple despike specifications are allowed. The despiking is done before the channel averaging (if any), so all channel numbers should be given in terms of the original SWARM channel range; 0 -> 16383. The despiking is very simple - the channels numbered between spike1 and spike2 (inclusive) are replaced by the average of the channels in the range left1 -> to left2 and right1 -> right2 (all ranges inclusive). Single channel ranges are allows (spike1 == spike2 is OK, etc.). Here's an example that removes two spikes in the RxB data for both sidebands of baseline 1-5, before reducing the resolution of all chunks by a factor of 4:
$ SMARechunker -r 4 -D '1-5:b:b:3:4740-4740:4730-4739:4741-4750' -D '1-5:b:b:3:3956-3961:3945-3955:3962-3970' \ -i ../../testFiles/180122_16:32:12/ -o by4despiked
In the example above, the first spike is a single channel spike at channel 4740, and the second spike is a wider spike covering channels 3956 through 3961.
The -f and -l optional switches allow you to specify the first and/or last scan to process. If you use one of these options the output dataset will have fewer scans than the input dataset. This can be useful for processing corrupted data sets..
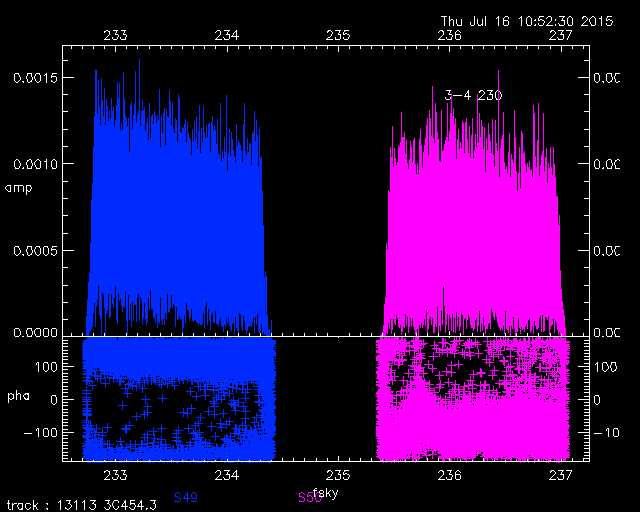
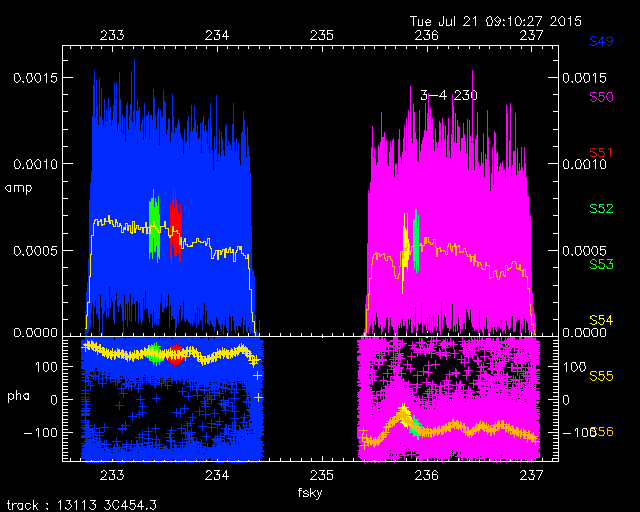
Here is another example. The left hand plot shows the raw USB data for chunks s49 and s50 on one baseline. The right hand plot shows the same dataset after processing by SMARechunker.
The command which produced the reduced resolution dataset plotted above was
$ SMARechunker -d -i ../../crap/150710_03:22:07/ -o testDir2 49:8000:9023:8 \ 50:11000:11511:8 49:6000:7023:8 50:12000:12511:8 49:0:16383:128 50:0:16383:128
The command above produces a dataset with the original SWARM chunks included (because of the -d) followed by a reduced resolution (by 8) copy of s49, channels 8000 through 9023 (49:8000:9023:8), followed by a reduced resolution copy of s50 channels 11000 through 11511 (50:11000:11511:8) followed by two more reduced bandwidth and resolution chunks, and ending with two chunks with the full bandwidth, but the resolution reduced by a factor of 128 (49:0:16383:128 50:0:16383:128). So the output dataset has chunks defined through s56.
If you just want to reduce the resolution of all SWARM chunks by the same factor, without trimming any channels off of the edges, then you can use the -r {n} option, where n is the number of SWARM channels to vector average in each SWARM chunk. If you use -r, you may not put any explicit chunk specifications on the command line. -r is only useful if you wish to reduce the resolution of all SWARM chunks by the same factor.
SMARechunker can process a 20GB dataset in about 1 minute (on the CF machine "jupiter"). Note that this program can take much longer to run if you are using a NFS cross-mounted filesytem. You may save time by copying the data directory to local storage on the machine you chose to run SMARechunker on.